| Introduction The translocation, t(10;11)(p12-p13;q14-q21), is a recurring abnormality observed in both acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML),1,2,3,4,5 and has been shown to involve fusion of AF10 on chromosome 10 with CALM on chromosome 11.6,7,8,9 AF10 was first identified as a fusion partner of MLL in a patient with a t(10;11)(p13;q23) diagnosed with AML-M5.10 Because MLL and AF10 are transcribed in opposite directions with respect to the centromere, fusions of MLL and AF10 often involve complex translocations and rearrangements in order to place the two genes in the appropriate orientation for the transcription of a functional chimeric gene. For this reason, it has been proposed that perhaps all t(10;11) observed in AML-M4 or AML-M5 involve AF10 and MLL and that the variability in assigned breakpoints is due to the complexity of the rearrangement.11,12 Subsequent to the identification of the MLL/AF10 fusion, AF10 was found to be translocated adjacent to a newly identified gene, CALM, in the monocytoid leukemia cell line, U937.6 CALM shares homology with the murine clathrin assembly protein ap-3; however, its function in normal human cells has yet to be determined. Recent studies have shown that CALM/AF10 fusions can be detected in bone marrow or peripheral blood samples from patients, the majority of which have a diagnosis of ALL or AML (AML-M0, or AML-M1).7,8,9,13 This has led to the conclusion that CALM and MLL rearrangements occur in morphologically distinct subsets of acute leukemia.8 In addition, based on the proposed functional structure of the CALM and AF10 genes, and the molecular breakpoints observed in U937, it has been proposed that it is the CALM/AF10 fusion product which is critical to the leukemic disease process.6 In order to investigate the role of AF10 and CALM in lymphoid and myeloid leukemia, we have used fluorescence in situ hybridization (FISH) and the reverse transcriptase polymerase chain reaction (RT-PCR) to study a series of nine patients cytogenetically characterized by the t(10;11)(p12-p13;q14-q23). The fusion of CALM and AF10 was detected in six patients including one with a diagnosis of AML-M5. A fusion of CALM and AF10 was not detected in the three remaining patient samples included in this study. Molecular characterization of the fusion gene product was possible for five of the six patient samples. In all cases, the CALM/AF10 gene product was detected. The reciprocal AF10/CALM product could only be detected in one of the five samples. We conclude that CALM/AF10 is a recurring abnormality in both lymphoid and myeloid leukemias of various types including AML-M5. In addition, due to the absent expression of the reciprocal AF10/CALM message, CALM/AF10 seems to be the fusion product critical to the disease state. Materials and methodsPatient samples The patient samples (peripheral blood, bone marrow aspirate or bone core biopsy) used in this study were determined previously by standard trypsin-Giemsa banding analysis to have rearrangements involving the short arm of chromosome 10 and the long arm of chromosome 11. All samples were obtained with the patient's informed consent. Those patients known to have rearrangements of MLL at 11q23 were excluded from this study. The samples used for RT-PCR were obtained at the time of diagnosis (patients 1, 2, 4, 5 and 9) or after treatment with chemotherapy (patients 3, 7 and 8). Samples were classified according to the French–American–British (FAB) criteria (Table 1). Fluorescence in situ hybridization The sequence-independent amplification (SIA) technique was used to prepare probes for the AF10 and CALM loci.14 Centre d'Etude du Polymorphisme Humain (CEPH) YAC 807b3 contains AF10 and CEPH YACs 785c1 and 914d9 flank the CALM gene. The primary SIA products from these YACs were provided by S.B. The probes were directly labeled with SpectrumGreen–dUTP or SpectrumOrange–dUTP (Vysis, Downers Grove, IL, USA) as previously described.15 For those patient samples for which material was available, dual color FISH was performed as described.15 CALM/AF10 RT-PCR assay Total RNA was isolated from cryopreserved patient material using the RNeasy RNA miniprep system (Qiagen, Valencia, CA, USA). Approximately 2 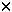 107 cells were used in each reaction. First-strand cDNA was made from approximately 5 107 cells were used in each reaction. First-strand cDNA was made from approximately 5 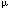 g RNA using an oligo dT primer and the Superscript preamplification system for first strand cDNA synthesis (GibcoBRL, Grand Island, NY, USA). The final product was diluted 1:2 with ddH2O and 1 g RNA using an oligo dT primer and the Superscript preamplification system for first strand cDNA synthesis (GibcoBRL, Grand Island, NY, USA). The final product was diluted 1:2 with ddH2O and 1 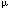 l was used as template for reverse transcriptase PCR using combinations of forward and reverse primers designed from the cDNA sequences of CALM and AF10 (Table 2). PCR amplification was performed in a 20 l was used as template for reverse transcriptase PCR using combinations of forward and reverse primers designed from the cDNA sequences of CALM and AF10 (Table 2). PCR amplification was performed in a 20 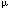 l reaction volume using 1 l reaction volume using 1 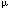 l of the diluted RT product, 200 l of the diluted RT product, 200 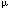 M each dNTP, 0.5 M each dNTP, 0.5 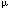 M each primer and 0.05 U Taq polymerase in a buffer containing 1.2 mM MgCl2 and 0.01% w/v gelatin. Reactions were carried out for 35 cycles of amplification at the following condition, 94°C for 1 min, 62°C for 1 min and 72°C for 1 min. PCR products were analyzed by electrophoresis on 1% agarose gels. M each primer and 0.05 U Taq polymerase in a buffer containing 1.2 mM MgCl2 and 0.01% w/v gelatin. Reactions were carried out for 35 cycles of amplification at the following condition, 94°C for 1 min, 62°C for 1 min and 72°C for 1 min. PCR products were analyzed by electrophoresis on 1% agarose gels. Subcloning and sequencing of PCR products PCR products were purified using QiaQuick columns (Qiagen) and sequenced directly using the dideoxy sequencing method and an ABI dRhodamine cycle sequencing kit (PE Applied Biosystems, Foster City, CA, USA). The sequences were resolved on an ABI Prism 377 DNA sequencer (Perkin Elmer, Norwalk, CT, USA). If multiple isoforms of the AF10/CALM fusion were observed in the amplification reaction, prior to sequencing, the purified products were cloned into the pCR II-TOPO cloning vector using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA). ResultsNine patient samples were included in this study. Six of the nine patients (patients 1–5 and 9) had a t(10;11)(p12-p13;q14-q21) in a bone marrow sample at the time of diagnosis. Pretreatment samples from patients 6 and 8 were not submitted for cytogenetic analysis, however, post-chemotherapy, residual disease samples were available. A bone marrow sample from patient 8 contained a complex t(10;11) involving a der(10)del(10)(p1?3p1?5)t(10;11)(p13;q23) and a der(11) t(10;11)(p13;q23). The t(10;11) was not present in a bone marrow sample taken from patient 7 at the time of initial diagnosis of CMMoL. However, the complex rearrangement described as, der(4)t(4;10)(q22;p13)t(10;11)(p13;q21), was observed in a later sample, taken after clinical progression to acute leukemia (AML-M4). Stored cells originally prepared for cytogenetic analysis were available for five of the nine patient samples (patients 1,2,5,6 and 8). Two color FISH with probes corresponding to CALM and AF10 on interphase and/or metaphase cells identified fusion products in four of the five samples (patients 1,2,5 and 6). In all four cases a single green (CALM) signal and single red (AF10) signal and two yellow (CALM/AF10, AF10/CALM) fusion signals were observed, indicating the presence of the normal chromosomes 10 and 11 as well as both derivative chromosomes. For patient 8, no metaphase cells containing the t(10;11) were observed upon hybridization with CALM and AF10 probes. Scoring of this hybridization using interphase cells was inconclusive because of high background. This patient presented with AML-M5, and at the time of cytogenetic analysis, had the clonal abnormality present in only two of 32 cells analyzed. Hybridization of MLL at that time revealed that MLL was present on the der(10) but not on the der(11). Frozen, viable cells were available for the preparation of RNA from eight of the nine patients. For five of the eight patients, RNA was made from the sample taken at the time of diagnosis (patients 1,2,4,5 and 9). For the remaining three patients, (patients 3,7 and 8), the samples were obtained after treatment with chemotherapy. Primers for the amplification of a normal AF10 transcript (NA.B628/AN.T288) and CALM transcript (NA.T501/AN.B497) were used to assess the quality of the RNA and the reverse transcriptase reaction. In all cases the normal AF10 and CALM gene products were successfully amplified. This is consistent with previous reports demonstrating expression of both CALM and AF10 in normal lymphocytes.6,11 Because it has been proposed that the fusion transcript critical to the leukemic state is the CALM/AF10 fusion, primer combinations specific for this product were used in an initial attempt to detect the aberrant message.6 CALM/AF10 fusion messages were detected in five of the eight RNA samples. Sequencing of the amplification products revealed four different CALM/AF10 fusion products from the five samples (Figure 1). The fusion of CALM and AF10 occurred at nucleotide position 2091 of the CALM cDNA and position 424 of AF10 in the fusion product amplified from patient 1 cDNA. CALM nucleotide 2091 was also the site of fusion in the CALM/AF10 transcript amplified from sample 2, however, in this case it was fused to AF10 nucleotide 883. Patients 3 and 4 had the identical fusion transcript, CALM2091/AF10979. Sample 5 involved a different CALM fusion site. In this sample CALM nucleotide 1926 was fused to AF10 nucleotide 589. The AF10/CALM reciprocal fusion product was only detected in RNA from patient 5. Three alternatively spliced transcripts were identified, AF10588/CALM1927, AF10588/CALM1987 and AF10588/CALM2092. CALM/AF10 fusion products were not detected in three of the patient samples included in our study (patients 7, 8 and 9). At the time of diagnosis of CMMoL, patient 7 had a gain of chromosome 8 as the sole clonal abnormality. The patient was treated with chemotherapeutic agents (mitoxantrone, Ara-C). One month later the disease progressed to AML-M4. Seven months following the initial induction chemotherapy, a t(10;11) occurred as the product of a complex rearrangement involving chromosomes 4, 10 and 11, (der(4)t(4;10) (q22;p13)t(10;11)(p13;q21)). A diagnosis sample was not available for patient 8, however, a post-chemotherapy (VP-16) sample revealed a complex rearrangement of chromosomes 10 and 11, (der(10)del(10)(p1?3p1?5)t(10;11) (p13;q23)). The third sample came from a patient (patient 9) diagnosed with T cell ALL and a balanced t(10;11)(p13;q14) as the sole abnormality in a pretreatment sample. DiscussionThis study describes the fusion of CALM and AF10 in six previously unreported patients with acute myeloid or lymphoid leukemia and the t(10;11)(p12–13;q14–21). All the patients with lymphoid leukemia had T cell ALL. Molecular characterization of the fusion sites using RT-PCR and sequence analysis was possible for five of the patient samples and resulted in the identification of four different fusion genes with varying breakpoints in both CALM and AF10. Three of the fusion transcripts isolated in this study are identical to those reported by others. CALM2091/AF10424 isolated here from a patient with AML-M0 is the same as the original CALM/AF10 fusion identified in the cell line U937.6 The CALM1926/AF10589 and CALM2091/AF10883 fusions were described in a recent study by Kumon et al.9 Not previously reported, is the CALM2091/AF10979 fusion transcript identified in two different patients in this study. This fusion transcript was also identified in the accompanying study by Bohlander et al.16 The CALM/AF10 fusion transcripts are characterized by four different breakpoints in AF10 and two breakpoints in CALM. Two of the AF10 breakpoints fall within the zinc finger region, resulting in a fusion gene predicted to contain a portion of this functional domain. In all cases the leucine zipper motif of AF10 is present in the CALM/AF10 fusion products. Both breakpoints in CALM are near the amino-terminus of the CALM gene, resulting in nearly the entire gene being present in the fusion product. This study supports the proposed idea that the CALM/AF10 fusion transcripts contain most of the functional domains of the two genes and, therefore, are critical to the disease process.6 Contrary to some other published studies, we were not able to detect the reciprocal AF10/CALM fusion products in the majority of our patient samples.6,9,13 This suggests that the AF10/CALM fusion is not critical to the disease process. Alternatively, it is possible that this fusion product is expressed at lower levels than the CALM/AF10 fusion product and was not detected by the methods used here. The AF10/CALM reciprocal fusion was successfully identified in one sample. In this case, three alternatively spliced AF10/CALM transcripts were detected. All three transcripts had the same AF10 breakpoint at nucleotide position 588. The various transcripts resulted from alternate splicing of two CALM exons bordered by nucleotide positions 1927 and 1986 and nucleotides 1987 and 2091. These alternatively spliced transcripts are consistent with CALM expression detected in non-malignant cells, and with the CALM exon-intron boundary sites predicted by the observed fusion transcripts.13 To date, including this study and the accompanying study by Bohlander et al, seven different CALM/AF10 fusion products have been described.6,9,13,16 Five different fusion transcripts have been detected in ALL and five in AML. CALM1926/AF10883, CALM2091/AF10589 and CALM2091/AF10979 have been observed in both AML and ALL. The CALM1926/AF10424 and CALM1926/AF10589 fusion transcripts have only been observed in ALL and the CALM2091/AF10424 and CALM2091/AF10883 transcripts in AML. It is of interest that all of the samples with ALL and the CALM/AF10 fusion express T cell-specific markers, indicating that there may be a specific association between CALM/AF10 fusion and T cell ALL. Somewhat unexpectedly, a fusion of CALM and AF10 was detected in a patient diagnosed with AML-M5. It has been proposed that CALM/AF10 fusions are limited to myeloblastic leukemias (AML-M0, AML-M1), whereas MLL/AF10 fusions are involved in leukemias with a monoblastic component (AML-M4, AML-M5).8,11 Patient 3 presented with a t(10;11)(p12;q21) as the sole abnormality at the time of diagnosis of AML-M5. The identification of a CALM/AF10 fusion in cases of AML-M5 is supported by the report of CALM/AF10 fusions in the two cell lines U937 and KP-MO-TS.6,9 Both of these cell lines were derived from patients with differentiated monocytic leukemia, AML-M5.17,18 Although much of the variability in assignment of cytogenetic breakpoints of the more differentiated leukemias may be a result of the complexity of MLL/AF10 rearrangements, the presence of a CALM/AF10 fusion, rather than the MLL/AF10 fusion, should be considered in patient samples with the t(10;11)(p12–13;q14–21) breakpoint. CALM/AF10 fusion products were not detected in three patients included in this study. One of the patients was diagnosed with AML-M5 and in another, the t(10;11) was only present after progression of CMMoL to AML-M4. Both of these cases involved complex chromosome rearrangements resulting in the t(10;11) and, therefore, are suggestive of an MLL/AF10 fusion product. Additionally, both samples were obtained after the patients had been treated with the topoisomerase II inhibitors, VP-16 and mitoxantrone. DNA-topoisomerase II targeting agents, are known to be associated with translocations involving MLL.19 The third sample for which we were unable to detect a CALM/AF10 fusion product came from a patient with T cell ALL and a balanced t(10;11)(p13;q14) as the sole abnormality in a pretreatment sample. It is possible that the CALM/AF10 fusion is present in this sample but in a copy number too low to be detected by the methods used in this study. This, however, is unlikely as cytogenetic analysis identified the clonal abnormality in five of 21 (25%) cells analyzed. Successful amplification of both the normal AF10 and CALM gene products suggest that the RNA isolated was of sufficiently high quality to detect large transcripts. It is possible that the breakpoints in CALM and/or AF10 fall outside the region tested in this study. Although all the CALM and AF10 breakpoints reported to date occur in the 3' end of CALM and the 5' end of AF10, it is possible that the site of fusion occurs more distally in CALM and/or proximally in AF10 in this patient. It is also possible that other genes are involved in this translocation. It is clear that CALM and AF10 play a critical role in the leukemogenic process as witnessed by the occurrence of both T cell lymphoid and myeloid leukemia with the t(10;11)(p12–p13;q14–q21) as a sole abnormality. Thus, this translocation should be added to the catalog of recurring rearrangements associated with T cell ALL. Continued study of these two genes and the CALM/AF10 fusion products will aid in our understanding of normal cellular processes as well as leukemogenesis. Acknowledgements
We are grateful to Marjorie Isaacson for retrieval of patient information and data management, and to Dr Carlos Suarez, Loyola University Medical Center, for referral of patient 5. This work was supported in part by the National Cancer Institute, CA 42557 (JDR). KC was supported by the G Harold and Leila Y Mathers Charitable Foundation.
|