| Introduction Over the last decade, molecular cytogenetic techniques such as fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH) have gained increasing importance in tumor cytogenetics.1,2 Nevertheless, standard chromosome analysis (CA) is still the method most frequently used to detect chromosomal aberrations in human cancer, especially in hematological malignancies.3,4 In lymphomas, CA may be hampered by difficulties in obtaining fresh lymph node samples and a sufficient number of high-quality metaphase spreads as well as by cytogenetic complexity. Interphase FISH can be used to identify primary changes like the t(14;18), the characteristic aberration of follicle center cell derived lymphomas (FCDL)5,6 as well as secondary changes like deletions in 6q that evolve during tumor progression and often determine the clinical course of the disease.7,8 Comparative genomic hybridization (CGH) might be helpful to detect secondary aberrations, if they involve gains and losses of genetic material.9,10 To date, there are two reports about the application of CGH in FCDL11,12 and several others that have compared the diagnostic power of CGH and CA to detect chromosomal imbalances in other lymphoma subtypes. However, with the exception of the study by Dierlamm et al,13 they were usually limited to the analysis of peripheral blood or bone marrow samples with a high portion of tumor cells.14,15,16,17 In this study, we present a systematic analysis to assess the reliability of CGH as compared to CA to detect gains and losses in lymph node specimens in FCDL. Materials and methodsMaterials Lymph node samples of 30 patients with histologically confirmed follicle center lymphoma (FCL) grade I/II (n = 24, centroblastic–centrocytic lymphoma according to Kiel classification) or with diffuse large cell lymphoma (n = 6, DLCL, centroblastic lymphoma according to Kiel classification) bearing the translocation t(14;18)(q32;q21) were selected for this study. The same cell suspensions were used for CGH, CA and FISH analyses. R banding analysis Metaphase spreads of tumor cells were prepared from short-term cultures of affected lymph nodes. Harvesting, slide preparation and R banding were performed as previously described.18 Karyotypes were described according to the International System for Human Genetic Nomenclature.19 Comparative genomic hybridization and digital image analysis Genomic DNA was prepared from fresh tumor tissue by using proteinase K digestion and phenol–chloroform extraction.20 CGH was performed according to standard methods.21 Briefly, normal human genomic DNA was labeled with digoxigenin-11-deoxyuridine triphosphate (Dig-dUTP; Boehringer, Mannheim, Germany) and tumor DNA was labeled with biotin-16-dUTP (Boehringer) by a standard nick translation reaction. One microgram of labeled tumor DNA, 1 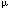 g of differentially labeled control DNA and 70 g of differentially labeled control DNA and 70 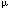 g of human Cot1-DNA (BRL Life Sciences, Gaithersburg, MD, USA) were cohybridized to metaphase cells prepared from peripheral blood of a healthy donor. After hybridization for 2 to 3 days and post-hybridization washes, control and test DNAs were detected via rhodamine and fluorescein isothiocyanate (FITC), respectively. Chromosomes were counterstained with 4,6-diamidino-2-phenylindole (DAPI). Images were acquired using an epifluorescence microscope (Axioplan Zeiss, Jena, Germany) connected with a commercially available image analysis system (ISIS; Metasystems, Sandhausen, Germany). Ratio profiles of the fluorescence intensities of tumor-DNAs and control-DNAs were calculated for each individual chromosome. For each patient, the mean ratio profiles of between 10 and 20 metaphases cells were computed and ratio values of 1.25 and 0.75 were defined as the upper and lower threshold for identification of chromosomal imbalances.22 g of human Cot1-DNA (BRL Life Sciences, Gaithersburg, MD, USA) were cohybridized to metaphase cells prepared from peripheral blood of a healthy donor. After hybridization for 2 to 3 days and post-hybridization washes, control and test DNAs were detected via rhodamine and fluorescein isothiocyanate (FITC), respectively. Chromosomes were counterstained with 4,6-diamidino-2-phenylindole (DAPI). Images were acquired using an epifluorescence microscope (Axioplan Zeiss, Jena, Germany) connected with a commercially available image analysis system (ISIS; Metasystems, Sandhausen, Germany). Ratio profiles of the fluorescence intensities of tumor-DNAs and control-DNAs were calculated for each individual chromosome. For each patient, the mean ratio profiles of between 10 and 20 metaphases cells were computed and ratio values of 1.25 and 0.75 were defined as the upper and lower threshold for identification of chromosomal imbalances.22 Interphase cytogenetics When both screening methods showed different results, discrepancies were further clarified, when possible, by interphase FISH using locus-specific probes. Fluorescence in situ hybridization (FISH) was performed on nuclei isolated from uncultured frozen lymph node biopsies as described recently,23 with slight differences depending on whether single or repetitive sequence probes and whether directly or indirectly labeled probes were used: cen7 (D7Z1; Oncor, Gaithersburg, MD, USA), cen12 (D12Z3; Vysis, Stuttgart, Germany), cen18 (D18Z1; Vysis), cenY (DYZ1; Vysis), 1p36 (D1Z2; Oncor), 6q21 (D6S301, YAC 882F8), 6q25 (D6S437, YAC 700H1), 8q24 (C-MYC, cosmids H4.1, P380J9 and MYC72), 10q23 (PTEN, YAC 746H8), 13q14 (RB1; Vysis), 16p13 (D16S3127, YAC 854E2) and 21q22 (D21S259; Vysis). Hybridization signals were analyzed by using a fluorescence microscope (Zeiss Axioskop, Oberkochen, Germany) and documented using the ISIS imaging system (Metasystems). In the mean 100 nuclei were evaluated per hybridization experiment. ResultsThe karyotypes, CGH and interphase FISH results are shown in Table 1. Conventional cytogenetics Among the 30 cases selected for the comparative analysis, 28 had chromosomal aberrations (93%), in one case no metaphases were obtained (case number 30) and in the other, the karyotype was normal (case number 12). The t(14;18)(q32;q21) translocation was present in 24 out of 30 cases (80%). In six cases of FCL grade I/II (Nos 6, 10, 12, 19, 21, 22) no t(14;18) could be identified with certainty. All cases with an abnormal karyotype had unbalanced aberrations (93%). One hundred and five imbalances were described (median 4, range 0–11), from which 69 were gains and 36 losses (66% vs 34%). The most recurrent secondary changes were total or partial gains on chromosomes 7 (10 cases: Nos 1, 5, 13, 14, 18, 19, 23, 24, 25, 29), 12 (seven cases: Nos 6, 13, 14, 15, 18, 23, 25), 18 (10 cases: Nos 2, 6, 7, 8, 10, 15, 16, 19, 28, 29) and X (nine cases: Nos 13, 15, 17, 18, 20, 21, 23, 24, 29). Frequent losses of chromosomal material were identified on chromosome arms 6q (seven cases: Nos 2, 18, 19, 21, 23, 27, 28) and 13q (five cases: Nos 16, 17, 20, 23, 25). Chromosomal material of unknown origin such as marker chromosomes (mar, four cases; 23, 26, 27 and 29), additions (add, eight cases; 18, 19, 20, 21, 25, 26, 27 and 28) and ring chromosomes (r, case 18) were reported in 34% of the cases with evaluable metaphases (10/29). Additionally, two cases presented incomplete karyotypes (25 and 27). Comparative genomic hybridization CGH analysis showed imbalances in 80% of cases (24/30). Fifty-six chromosomal imbalances were identified (median 1, range 0–7), from which 40 were gains and 16 losses (71% vs 29%). As by CA, the most frequent gains were identified on chromosomes 7 (nine cases: Nos 1, 13, 14, 18, 20, 23, 24, 25, 27), 12 (four cases: Nos 14, 15, 18, 23), 18 (six cases: Nos 2, 7, 8, 10, 25, 28) and X (seven cases: Nos 13, 15, 17, 18, 23, 24, 26), whereas the most frequent losses mapped to the long arms of chromosome 6 (five cases: Nos 2, 18, 23, 24, 28) and 13 (two cases: Nos 17, 23). Comparison between R banding and CGH data We have divided the patients into two different subgroups: those with completely resolved karyotypes after CA and those with partially unresolved chromosome aberrations after CA. Altogether, in six patients identical results were obtained by both methods (Nos 1, 2, 3, 10, 11, 23). The other 24 patients presented one or more discrepancies. We found 73 discrepant results, 11 were detected only by CGH and 62 only by R banding. CA detected significantly more imbalances than CGH (Wilcoxon test, P < 0.0005). Figure 1 shows the ideograms with all the discrepancies found as well as the common findings. Comparison of CA and CGH on patients with completely resolved karyotype These 17 cases represent the ideal way to compare both techniques. In five patients (1, 2, 3, 10 and 11), CA and CGH gave identical results. In 12 patients, 22 discrepant findings (median 1, range 0–5) were found. All but one were identified by CA, eight of them in cases in which no imbalances were seen by CGH. Nine discrepant alterations were subjected to interphase FISH validation. Four imbalances detected only by CA were confirmed. Four imbalances detected only by CA and one imbalance detected only by CGH could not be confirmed. It cannot be ruled out, however, that the FISH probes hybridized to loci outside the gained or lost regions. Figure 2 shows the CGH profile of patient No. 13 with its respective R-banding karyotype. In this patient, the gain of 12p observed by CA was considered not significant by CGH for being in the limit of detection. Comparison of CA and CGH on patients with partially unresolved karyotype Fifty-one discrepancies were found in this group (70%, median 4, range 0–11). The number of discrepancies was significantly higher (Mann–Whitney U test, P = 0.003) than in patients with completely resolved karyotypes. Except for patient 23, in whom the same imbalances were found, all the remaining cases presented discrepancies. Forty-one imbalances were identified by CA, 10 by CGH. Of course, we cannot rule out that due to the complexity of the karyotypes chromosomal imbalances described by CA indeed represent balanced aberrations, e.g. in case 27, several additions and deletions, as well as a marker chromosome were reported, so perhaps some of those aberrations taken as imbalances are balanced rearrangements. By FISH, 13 imbalances detected only by CA and two imbalances detected only by CGH were confirmed. One imbalance each detected only by CA and by CGH, respectively, could not be confirmed. As can be seen from Table 2, CA was able to identify significantly more chromosomal imbalances in FCL and DLCL than CGH, both in the group of patients with completely resolved karyotypes and in the group with partially unresolved karyotypes. DiscussionFollicle center cell derived lymphomas are cytogenetically characterized by the presence of the t(14;18) translocation. Since this primary immortalizing event does not render the cells malignant, it is thought that additional secondary aberrations are necessary for tumorigenesis. In FCDL, the most frequently reported secondary changes are gains of chromosomes 7, 12, 18 or der(18), and X; as well as loss of chromosome 6q.11,12,24 In our study, both CGH and CA showed that the most recurrent imbalances associated with the t(14;18) were in line with previous reports. Different reasons may be responsible for the higher sensitivity of CA compared to CGH and for the discrepancies found. Most importantly, FCL may contain less than 50% tumor cells which is below the diagnostic threshold of CGH.21 The same holds true for subclones that often represent small proportions of tumor cells, albeit they may be important for the progression of the disease.24 Balanced rearrangements, which were present in a high number in addition to the characteristic t(14;18) in the FCDL studied, as well as tri- or tetraploid clones are not detectable by CGH.25 Moreover, some chromosomal regions, e.g. 1ptel, 16p, 19 and Y, give sometimes inconsistent hybridization results, which should be interpreted with care.9 Otherwise, chromosome aberrations may remain undetected by CA because of a low spontaneous proliferation rate of the tumor cells and because of an insufficient banding quality or complex chromosome rearrangements which hinder a full resolution of the karyotype. Discrepant findings might also result from different percentages of tumor cells or various subclones in portions of the lymph nodes that were divided to be analyzed by CA and CGH. We tried to rule this out by using the same cell suspensions for both methods. In conclusion, CGH can be considered a useful method to detect secondary aberrations in FCDL when no metaphases are found or when no fresh material is available, however, for routine diagnosis, CA is still the gold standard. For research purposes, CGH and CA should be combined to obtain a deeper insight into chromosomal imbalances. Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (Grant Be1454/5–2), the Deutsche Krebshilfe (Grant M 47/95 Be I and Grant 10–1556 Schl4), the Interdisciplinary Center for Clinical Cancer Research of the University of Kiel and the Hermann and Lilly Schilling-Foundation. We would like to thank Maria Jose Calasanz for her critical review of the manuscript and helpful advice and A Borowski, A Schneider, F Kahlert for excellent technical assistance.
|